HKUST’s Meta-analysis Shows SARS-CoV-2 Variants Unlikely to Affect T Cell Responses
In a new study, scientists at the Hong Kong University of Science and Technology (HKUST) have revealed that most T cell epitopes known to be targeted upon natural infection are seemingly unaffected by current SARS-CoV-2 variants.
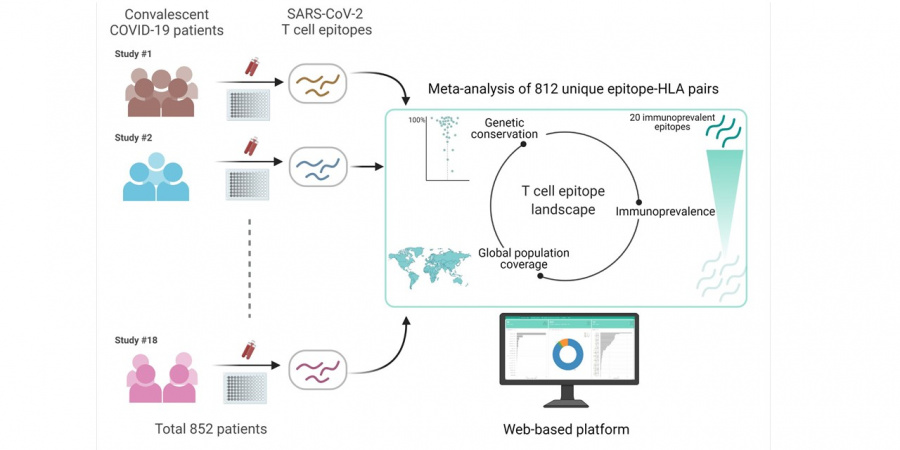
In their latest research, the team compiled and analysed data from 18 immunological studies of T cell responses involving over 850 recovered COVID-19 patients from across four continents who are well-distributed in age, gender, disease severity and blood collection time. They demonstrated that T cells in these patients targeted fragments (epitopes) of almost all of the virus’ proteins, including the spike protein that is a main target of many existing vaccines. As an important finding, based on analysis of over 850,000 SARS-CoV-2 genetic sequences from around the world, most of these epitopes appeared to be unaffected by the current variants of concern.
“We focused specifically on recovered COVID-19 patients as their immune responses are representative of effective responses against the virus,” said Prof. Ahmed Abdul QUADEER, Research Assistant Professor from the Department of Electronic and Computer Engineering, who is the first author of the research.
“We believe this is good news, particularly for vaccines. In contrast to antibody responses which have been shown to be affected by variants, our analysis would suggest T cell responses may be relatively robust, assuming that vaccine responses mimic those of natural infection,” said Prof. Matthew MCKAY from the Departments of Electronic & Computer Engineering and Chemical & Biological Engineering, who co-led the research.
While revealing more than 700 T cell epitopes, the team’s analysis identified 20 specific epitopes – which they refer to as immunoprevalent – that induced T cell responses in multiple independent cohorts and in a large fraction of tested patients. Of these, five appeared highly immunoprevalent, registering T cell responses in at least four separate immunological studies.
“The identified immunoprevalent epitopes appear to represent parts of the virus that are commonly targeted by T cells in recovered COVID-19 patients,” added Prof. McKay. “It is possible, but subject to further experimental investigation, that targeting these epitopes may play a role in contributing to favourable disease outcomes.”
The compiled data has been integrated into a web platform, which the team plans to keep updated as more immunological studies of SARS-CoV-2 T cell epitopes are reported.
“The platform is an online resource for the global research community and complements ongoing research on understanding T cell responses against SARS-CoV-2. It serves to guide studies related to COVID-19 vaccines and diagnostics,” added Prof. Quadeer.
Their findings have just been published in the scientific journal Cell Reports Medicine.
(This news was originally published on EurekAlert on May 26, 2021.)