HKUST Breakthrough Identifies Rare Tumor Cell “Spies” Shedding Light on Previously Hidden Cancer Features
Researchers at the Hong Kong University of Science and Technology (HKUST) developed a novel technology which allows genomic DNA and RNA sequencing to be carried out simultaneously in single cells of both frozen and fresh tissues, and identified rare brain tumor cell “spies” disguised as normal cells with this method. This breakthrough facilitates cancer research for some of the most complex and rare tumors, opening new directions for drug target discovery in the future.
Genomic DNA and RNA sequencing is crucial for determining the treatment for cancer, as it offers important information on the tumor’s genomic and molecular composition, or cellular heterogeneity, which influences the disease pathology as well as the tumor’s ability to develop drug resistance. Our present knowledge about cancers does not fully explain why tumors relapse or become resistant to treatment; exploring new dimensions of the tumor composition at high resolution by looking at the DNA and RNA together may provide answers. Existing technologies, however, have limited applicability to simultaneously perform DNA and RNA sequencing in single cells from frozen biobanked tissues, yet these frozen tissues make up most of the readily available clinical cancer samples.
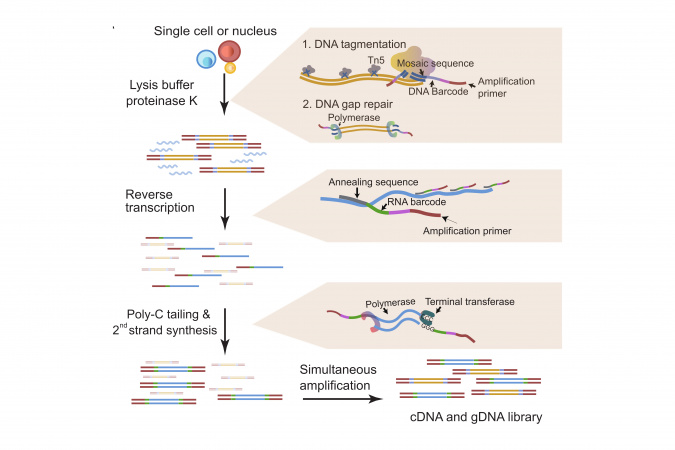
Now, a team led by Prof. Angela WU, Associate Professor of HKUST’s Division of Life Science and Department of Chemical and Biological Engineering, and her postdoctoral fellow Dr. YU Lei developed a new versatile single-cell multi-omic profiling technology scONE-seq, which can analyze frozen cells and difficult-to-obtain cell types like bone and brain. This new method can also simultaneously collect genomic and transcriptomic information in a tumor through a one-pot reaction.
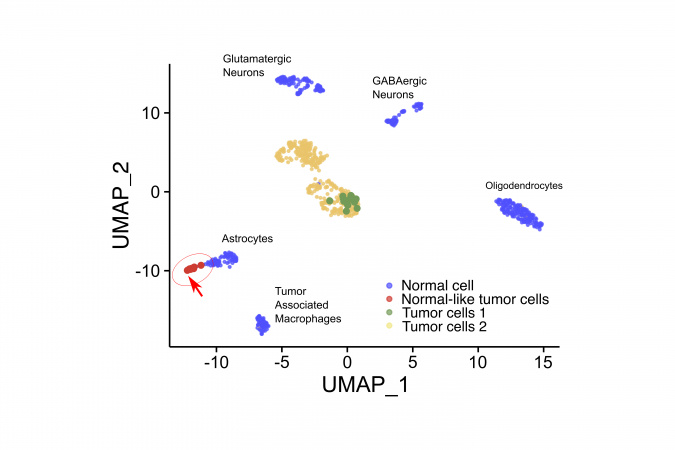
Astrocytoma is a deadly and aggressive type of brain tumor, and patients with this type of tumor have a survival rate of only around 5% within five years of diagnosing the disease. Using their new single-cell technology, the team has discovered a small and unique tumor cell subpopulation in a patient’s astrocytoma sample. This unique tumor population disguised themselves as normal astrocytes of the brain, which could escape detection using other common tumor sequencing methods. In addition, this “spy” tumor cell also showed molecular features that are related to drug resistance. The comprehensive role of this “spy” tumor cell in tumor progression will be an important direction for future investigations of this disease and possible drug targets.
Prof. Angela Wu said, “By identifying rare tumor cells which might be missed by previous approaches and result in failure to respond to therapy, the scONE-seq approach represents a new path to discovering drug targets and the development of new drugs. We plan to continue our work, using scONE-seq to profile a larger patient cohort, and hope to have more clinically translational outcomes in the future.”
This study was done in collaboration with Prof. WANG Jiguang and his team from HKUST’s Division of Life Science and Department of Chemical and Biological Engineering, as well as clinician scientists Dr. Danny CHAN, Dr. Aden CHEN, Dr. NG Ho-Keung, and Dr. POON Wai-Sang at the Chinese University of Hong Kong and Prince of Wales Hospital. The discovery was recently published in Science Advances.
(This news was originally published by the HKUST Public Affairs Office here.)